This tutorial is the second part of the introduction to simple linear regression in R, the use of ANOVAs with categorical predictors.
First we’re going to load in all the packages we’ll be using in this analysis.
library(agridat) # a package of agricultural datasets
library(summarytools) # useful functions for summarising datasets
library(dplyr) # manipulating data
library(ggplot2) # plotting
library(gridExtra) # plotting in panels
library(car) # extensions for regression
library(AICcmodavg) # calculates AICcAs we’re plotting with ggplot I’m going to set the theme now for the whole tutorial to make it look nicer to me - see the plotting tutorial for more information on the plotting commands.
theme_set(theme_bw() + theme(panel.grid = element_blank()))We’re going to be using the apples data from the agridat package, an experiment from the 1980s on the impact of spacing, root stock and cultivars on apple tree yields. We’ll be focusing on the impact of spacing, which came in three different treatments: 6m, 10m and 14m apart.
apples <- agridat::archbold.apple
head(apples)## rep row pos spacing stock gen yield trt
## 1 R1 2 1 6 Seedling Redspur 70.9 601
## 2 R1 2 2 6 Seedling Golden 130.9 602
## 3 R1 2 8 6 MM111 Redspur 114.5 611
## 4 R1 2 7 6 MM111 Golden 90.5 612
## 5 R1 2 3 6 M0007 Redspur 151.8 671
## 6 R1 2 4 6 M0007 Golden 125.0 672str(apples)## 'data.frame': 120 obs. of 8 variables:
## $ rep : Factor w/ 5 levels "R1","R2","R3",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ row : int 2 2 2 2 2 2 2 2 2 2 ...
## $ pos : int 1 2 8 7 3 4 5 6 16 12 ...
## $ spacing: int 6 6 6 6 6 6 6 6 10 10 ...
## $ stock : Factor w/ 4 levels "M0007","MM106",..: 4 4 3 3 1 1 2 2 3 1 ...
## $ gen : Factor w/ 2 levels "Golden","Redspur": 2 1 2 1 2 1 2 1 2 2 ...
## $ yield : num 70.9 130.9 114.5 90.5 151.8 ...
## $ trt : int 601 602 611 612 671 672 661 662 1011 1071 ...print(dfSummary(apples), method = "render")Data Frame Summary
apples
Dimensions: 120 x 8Duplicates: 0
| No | Variable | Stats / Values | Freqs (% of Valid) | Graph | Valid | Missing | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rep [factor] | 1. R1 2. R2 3. R3 4. R4 5. R5 |
|
120 (100%) | 0 (0%) | |||||||||||||||||||||
| 2 | row [integer] | Mean (sd) : 9 (4.4) min < med < max: 2 < 9 < 16 IQR (CV) : 7.2 (0.5) | 15 distinct values | 120 (100%) | 0 (0%) | |||||||||||||||||||||
| 3 | pos [integer] | Mean (sd) : 9.2 (4.9) min < med < max: 1 < 10 < 17 IQR (CV) : 9 (0.5) | 17 distinct values | 120 (100%) | 0 (0%) | |||||||||||||||||||||
| 4 | spacing [integer] | Mean (sd) : 10 (3.3) min < med < max: 6 < 10 < 14 IQR (CV) : 8 (0.3) |
|
120 (100%) | 0 (0%) | |||||||||||||||||||||
| 5 | stock [factor] | 1. M0007 2. MM106 3. MM111 4. Seedling |
|
120 (100%) | 0 (0%) | |||||||||||||||||||||
| 6 | gen [factor] | 1. Golden 2. Redspur |
|
120 (100%) | 0 (0%) | |||||||||||||||||||||
| 7 | yield [numeric] | Mean (sd) : 145.4 (47.5) min < med < max: 64.1 < 147.1 < 282.3 IQR (CV) : 68.3 (0.3) | 85 distinct values | 92 (76.67%) | 28 (23.33%) | |||||||||||||||||||||
| 8 | trt [integer] | Mean (sd) : 1036.5 (329.4) min < med < max: 601 < 1036.5 < 1472 IQR (CV) : 735.5 (0.3) | 24 distinct values | 120 (100%) | 0 (0%) |
Generated by summarytools 0.9.3 (R version 3.6.0)
2019-05-31
So do apples that are closer together have lower yield? There are only 3 spacing values so we’ll convert it to a categorical variable using as.factor.
apples$spacing2 <- as.factor(apples$spacing)
ggplot(apples, aes(spacing2, yield)) +
geom_boxplot() +
labs(x = "Spacing (m)", y = "Yield (kg)")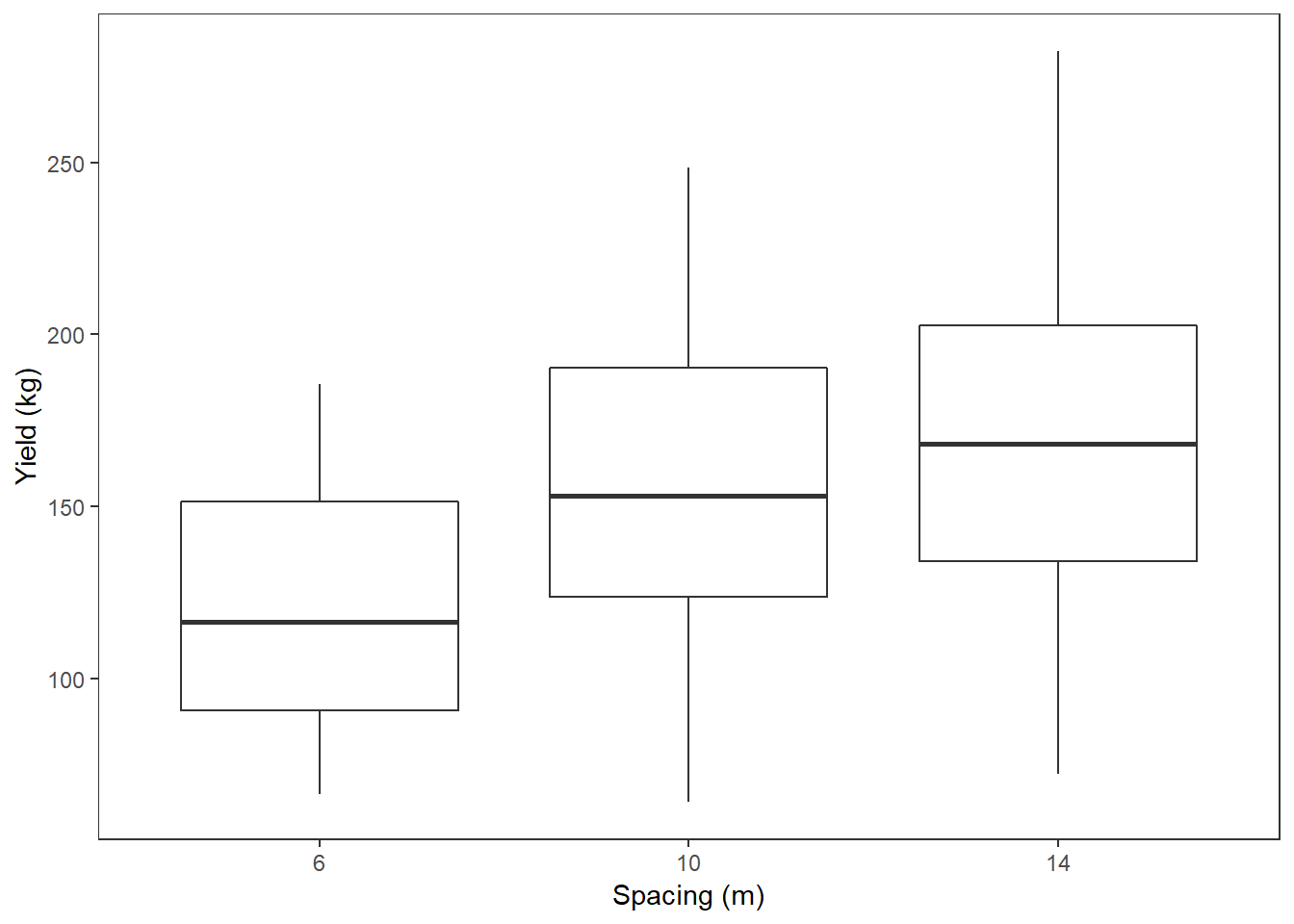
Within R lm and aov give the same result with a single categorical predictor only difference is output of summary functions, which can be changed using summary.lm and summary.aov
now run linear model with lm to see if difference is statistically significant
apples.m <- lm(yield ~ spacing2, data = apples)
summary(apples.m)##
## Call:
## lm(formula = yield ~ spacing2, data = apples)
##
## Residuals:
## Min 1Q Median 3Q Max
## -92.389 -30.577 -3.516 33.192 117.628
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 120.566 7.382 16.332 < 2e-16 ***
## spacing210 35.924 11.073 3.244 0.001659 **
## spacing214 44.107 10.966 4.022 0.000121 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 43.67 on 89 degrees of freedom
## (28 observations deleted due to missingness)
## Multiple R-squared: 0.1742, Adjusted R-squared: 0.1556
## F-statistic: 9.385 on 2 and 89 DF, p-value: 0.0002003output gives the difference between the 10m spacing and the 6m spacing, and the 14m spacing and the 6m spacing. The 6m spacing is given by the intercept
use plots to examine residuals.
par(mfrow=c(2,2));plot(apples.m);par(mfrow=c(1,1)) 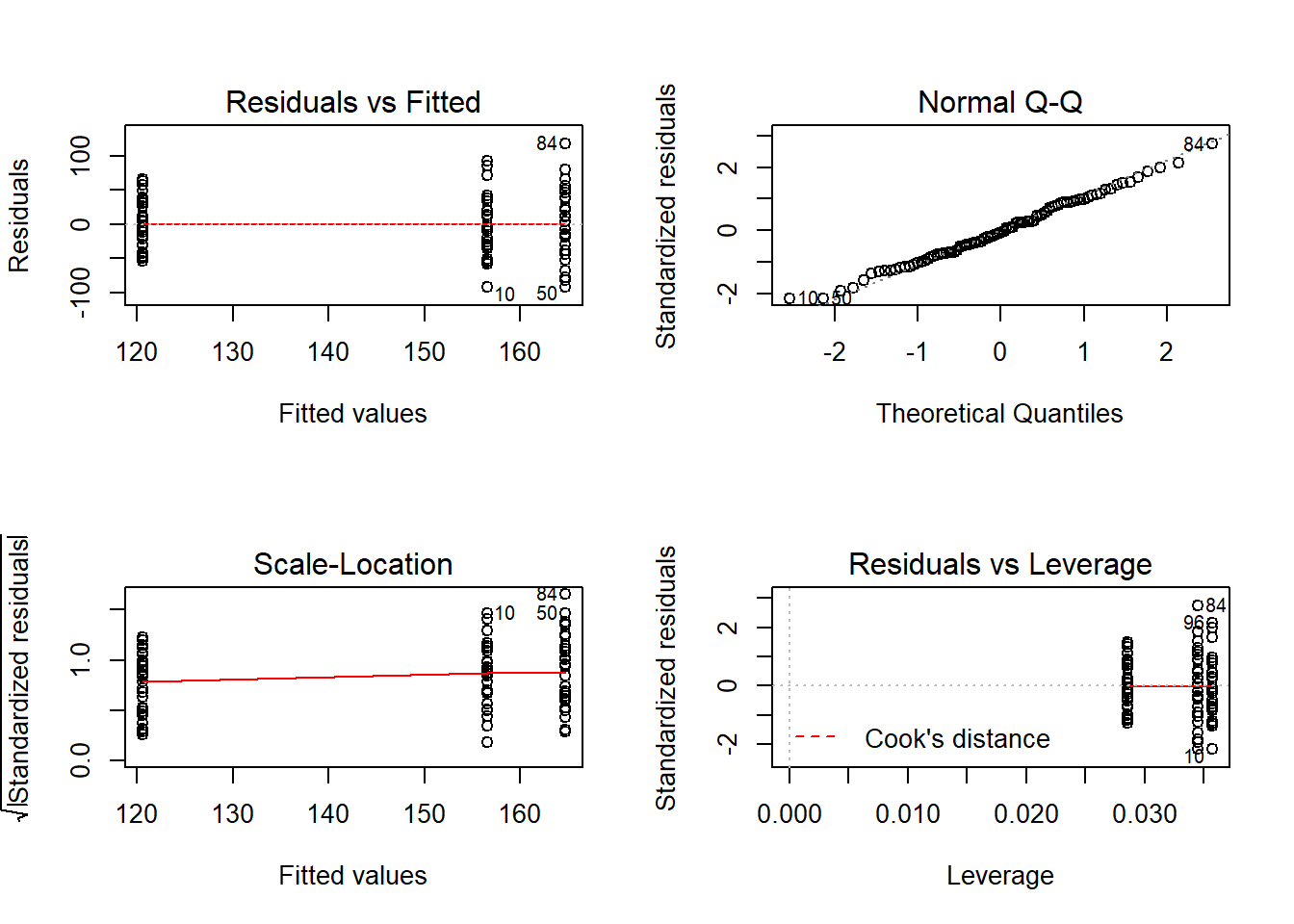
qqp(apples.m)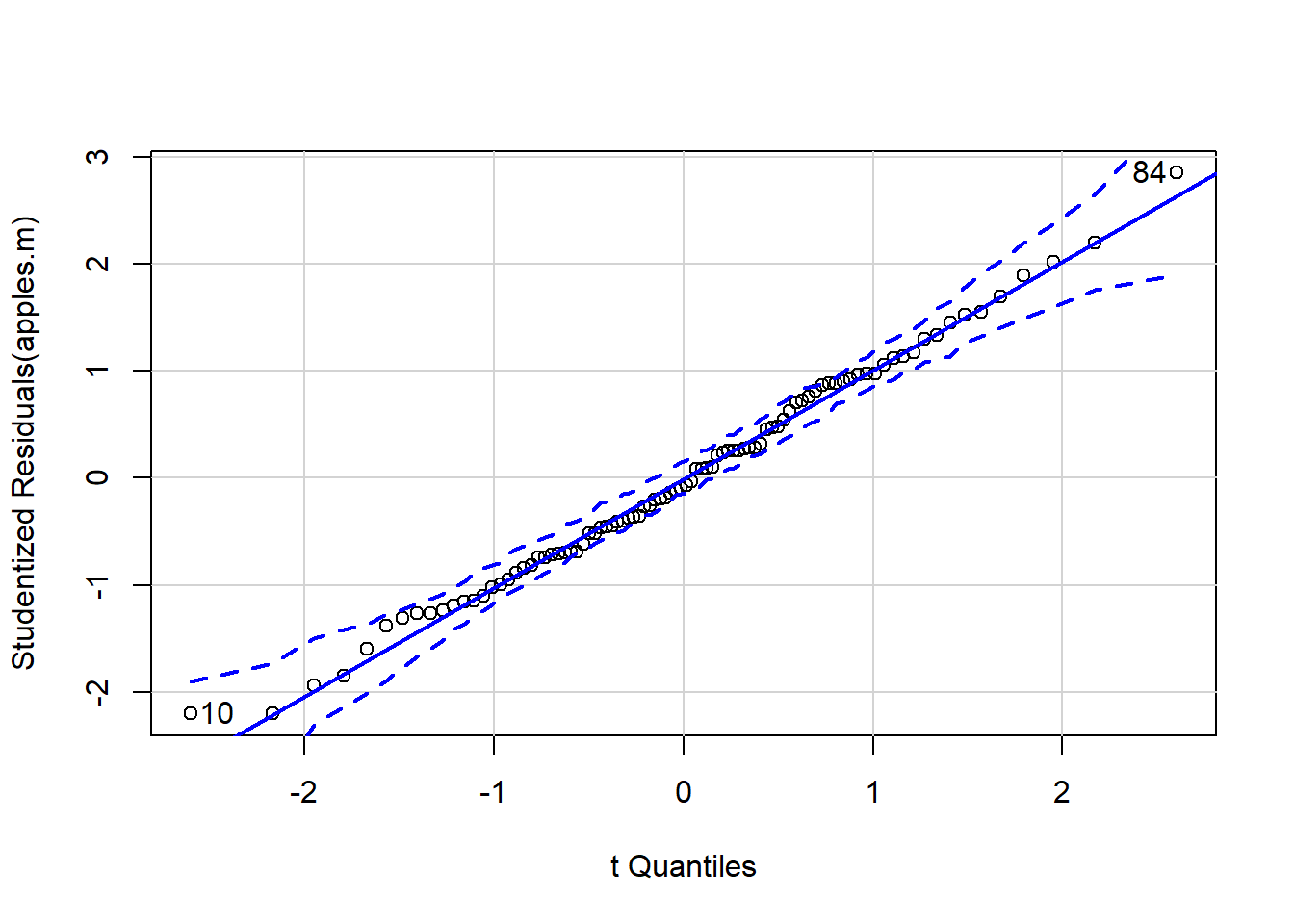
## [1] 10 84If wish to do a Tukey’s HSD post-hoc test then need to fit model with aov rather than lm
apples.aov <- aov(yield ~ spacing2, data = apples)
summary(apples.aov)## Df Sum Sq Mean Sq F value Pr(>F)
## spacing2 2 35801 17900 9.385 2e-04 ***
## Residuals 89 169750 1907
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## 28 observations deleted due to missingnesssummary(apples.m) # compare to lm output - different format but same numbers##
## Call:
## lm(formula = yield ~ spacing2, data = apples)
##
## Residuals:
## Min 1Q Median 3Q Max
## -92.389 -30.577 -3.516 33.192 117.628
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 120.566 7.382 16.332 < 2e-16 ***
## spacing210 35.924 11.073 3.244 0.001659 **
## spacing214 44.107 10.966 4.022 0.000121 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 43.67 on 89 degrees of freedom
## (28 observations deleted due to missingness)
## Multiple R-squared: 0.1742, Adjusted R-squared: 0.1556
## F-statistic: 9.385 on 2 and 89 DF, p-value: 0.0002003summary.lm(apples.aov) # can get equivalent output to lm using summary.lm##
## Call:
## aov(formula = yield ~ spacing2, data = apples)
##
## Residuals:
## Min 1Q Median 3Q Max
## -92.389 -30.577 -3.516 33.192 117.628
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 120.566 7.382 16.332 < 2e-16 ***
## spacing210 35.924 11.073 3.244 0.001659 **
## spacing214 44.107 10.966 4.022 0.000121 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 43.67 on 89 degrees of freedom
## (28 observations deleted due to missingness)
## Multiple R-squared: 0.1742, Adjusted R-squared: 0.1556
## F-statistic: 9.385 on 2 and 89 DF, p-value: 0.0002003TukeyHSD(apples.aov)## Tukey multiple comparisons of means
## 95% family-wise confidence level
##
## Fit: aov(formula = yield ~ spacing2, data = apples)
##
## $spacing2
## diff lwr upr p adj
## 10-6 35.923571 9.530402 62.31674 0.0046832
## 14-6 44.106700 17.967561 70.24584 0.0003526
## 14-10 8.183128 -19.396838 35.76309 0.7598733ANOVA with unbalanced designs and > 1 predictor
Balanced designs have the same number of reps per treatment. This is often not the case, and the study is unbalanced. If the study is unbalanced the way in which predictors are evaluated impacts the result. The different ways of evaluating predictors are known as Type I, Type II and Type III ANOVAs
To find out more about this go to http://goanna.cs.rmit.edu.au/~fscholer/anova.php
For type III tests to be correct need to change the way R codes factors
options(contrasts = c("contr.sum", "contr.poly")) This won’t change type I or II Default is: options(contrasts = c(“contr.treatment”, “contr.poly”))
A = c("a", "a", "a", "a", "b", "b", "b", "b", "b", "b", "b", "b")
B = c("x", "y", "x", "y", "x", "y", "x", "y", "x", "x", "x", "x")
C = c("l", "l", "m", "m", "l", "l", "m", "m", "l", "l", "l", "l")
response = c( 14, 30, 15, 35, 50, 51, 30, 32, 51, 55, 53, 55)model = lm(response ~ A + B + C + A:B + A:C + B:C)
anova(model) # Type I tests## Analysis of Variance Table
##
## Response: response
## Df Sum Sq Mean Sq F value Pr(>F)
## A 1 1488.37 1488.37 357.6869 7.614e-06 ***
## B 1 18.22 18.22 4.3798 0.0905765 .
## C 1 390.15 390.15 93.7610 0.0001995 ***
## A:B 1 278.68 278.68 66.9722 0.0004431 ***
## A:C 1 339.51 339.51 81.5909 0.0002778 ***
## B:C 1 8.51 8.51 2.0444 0.2121592
## Residuals 5 20.81 4.16
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Anova(model, type="II") # Type II tests using car package## Anova Table (Type II tests)
##
## Response: response
## Sum Sq Df F value Pr(>F)
## A 1022.01 1 245.6103 1.923e-05 ***
## B 131.25 1 31.5421 0.0024764 **
## C 278.68 1 66.9722 0.0004431 ***
## A:B 180.01 1 43.2593 0.0012194 **
## A:C 321.01 1 77.1445 0.0003174 ***
## B:C 8.51 1 2.0444 0.2121592
## Residuals 20.81 5
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Anova(model, type="III") # Type III tests using car package## Anova Table (Type III tests)
##
## Response: response
## Sum Sq Df F value Pr(>F)
## (Intercept) 9490.0 1 2280.6425 7.605e-08 ***
## A 724.5 1 174.1138 4.465e-05 ***
## B 184.5 1 44.3408 0.0011526 **
## C 180.0 1 43.2593 0.0012194 **
## A:B 180.0 1 43.2593 0.0012194 **
## A:C 321.0 1 77.1445 0.0003174 ***
## B:C 8.5 1 2.0444 0.2121592
## Residuals 20.8 5
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Balanced design shows no difference between type I, II and III
A = c("a", "a", "a", "a", "a", "a", "b", "b", "b", "b", "b", "b")
B = c("x", "y", "x", "y", "x", "y", "x", "y", "x", "y", "x", "y")
C = c("l", "l", "m", "m", "l", "l", "m", "m", "l", "l", "m", "m")
response = c( 14, 30, 15, 35, 50, 51, 30, 32, 51, 55, 53, 55)model = lm(response ~ A + B + C + A:B + A:C + B:C)
anova(model) # Type I tests## Analysis of Variance Table
##
## Response: response
## Df Sum Sq Mean Sq F value Pr(>F)
## A 1 546.75 546.75 1.9146 0.2250
## B 1 168.75 168.75 0.5909 0.4768
## C 1 315.38 315.38 1.1044 0.3414
## A:B 1 70.08 70.08 0.2454 0.6413
## A:C 1 0.38 0.38 0.0013 0.9725
## B:C 1 15.04 15.04 0.0527 0.8276
## Residuals 5 1427.87 285.57Anova(model, type="II") # Type II tests## Anova Table (Type II tests)
##
## Response: response
## Sum Sq Df F value Pr(>F)
## A 782.04 1 2.7385 0.1589
## B 168.75 1 0.5909 0.4768
## C 315.38 1 1.1044 0.3414
## A:B 84.37 1 0.2955 0.6101
## A:C 0.38 1 0.0013 0.9725
## B:C 15.04 1 0.0527 0.8276
## Residuals 1427.87 5Anova(model, type="III") # Type III tests ## Anova Table (Type III tests)
##
## Response: response
## Sum Sq Df F value Pr(>F)
## (Intercept) 16380.4 1 57.3593 0.0006367 ***
## A 782.0 1 2.7385 0.1588624
## B 168.7 1 0.5909 0.4767865
## C 315.4 1 1.1044 0.3414277
## A:B 84.4 1 0.2955 0.6100929
## A:C 0.4 1 0.0013 0.9724954
## B:C 15.0 1 0.0527 0.8275710
## Residuals 1427.9 5
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The choice between the types of ANOVAs is beyond the scope of this R club and is hypothesis dependent.